July 27, 2012 Aipotu wins Science
Prize for Inquiry-based Instruction
May 24, 2017 Aipotu updated for new OS X security.
May 27, 2021 jsAipotu: a version of Aipotu that runs in a browser released for
testing.
Aipotu
Pronounced “ā poh too”. The name is “Utopia” reversed: the software creates a
utopia that simulates the genetics, biochemistry, molecular biology, and evolution of organisms in
a biologically reasonable and pedagogically relevant way.
Students apply the tools of four of the major disciplines in modern biology (genetics, biochemistry,
molecular biology, and evolution) to develop an integrated understanding of the formation of color
in a simulated flower. They do this by using four Workbenches, one for each discipline.
Students can save interesting organisms to the Greenhouse that communicates among
the Workbenches.
The four individual Workbenches in Aipotu can be used individually to explore an individual
topic. In addition, they can be used in any order to explore flower color.
The software is available for free as are sample lab manuals and other supporting materials. There is a users'
group where teachers can share ideas, ask questions, and develop materials using Aipotu.
The software and teaching materials were developed as a multi-year collaboration that included:
- Brian White of the University of
Massachusetts, Boston Biology Department
- Ethan Bolker of the University of
Massachusetts, Boston Math & Computer Science Departments
- A large group of students in the University of Massachusetts, Boston Computer Science Department
- The students and Graduate Teaching Assistants who have used Aipotu in the classroom.
This software and teaching materials have been used by more than 2,000 students at UMB and
elsewhere.
I have prepared a short handout describing Aipotu. It is available by clicking the image
to the right.
Overview
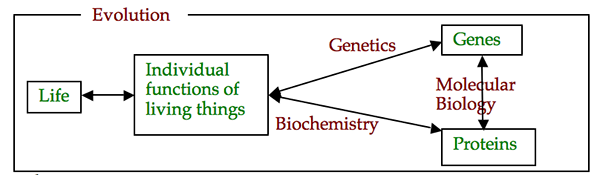
Aipotu is a BioQUEST software simulation that integrates genetics, biochemistry, molecular biology, and evolution to study a biological phenomenon. It is designed to show students the connections between these three key disciplines of modern molecular genetics. It is based on "Botstein's Triangle"; (Science 240:1439 (1988)) shown below:
That is:
- Life can be explained in terms of The Individual Functions of Living Things.
- Genetics explains The Individual Functions of Living Things in terms of Genes.
- Biochemistry explains The Individual Functions of Living Things in terms of Proteins.
- Molecular Biology connects Genetics and Biochemistry by explaining how Genes encode Proteins.
- Evolution provides a context that explains how these features changed over time to become what they are.
- The phenomenon under study - the Individual function of this living thing - is the color of a simulated flower (see below).
The student is provided with a starting set of flowers in the Greenhouse; these organisms are saved as organism files, each of which contains two DNA sequences. These organisms are loaded into the Greenhouse when the program starts.
Aipotu is available in two versions:
- Aipotu stand-alone application.
- This is available for download from the Download link at the left.
- It requires java to run.
- It has all the features described in the documentation.
- jsAipotu a browser-based web app.
- This is accessible from this link.
- No download or install is required
- Java is not required
- It works best in Google Chrome and Firefox
- It automatically saves the Greenhouse in your browser's local storage. This storage is retained even
when you reload the page but it is not transferrable to another browser or computer. You
can also save the Greenhouse to a file that you can move to another computer.
- It has all the core features of the stand-alone application, EXCEPT:
-
- It cannot save images.
- You can enable or disable mutations in Evolution but you cannot change mutation frequencies
- In Evolution, you must click each time you want to run a generation.
- Evolution and Mutation run a little slower than with the stand-alone application.
- The layout isn't quite as pretty as the stand-alone app
Genetics
In the Genetics Workbench, students use the tools of genetics to determine the alleles present
in the Greenhouse strains and their interactions.
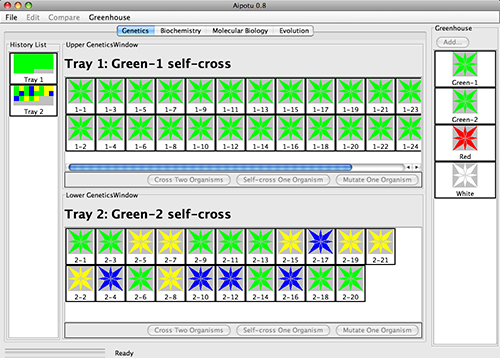
Students can:
- Cross any two organisms.
- Self-cross any organism.
- Subject any organism to random mutagenesis.
- Save any organism to the Greenhouse (right side) for analysis in the other Workbenches.
A record of each cross is preserved in the History List on the left side. Double-clicking an entry in the History List allows it to be brought to the main window so the organisms in it can be crossed.
Algorithm: All aipotian organisms are diploid - they contain two DNA sequences encoding pigment
proteins. In a cross or self-cross, offspring are generated by random choice of parental alleles. In
mutagenesis, offspring are cloned from the starting organism and their DNA sequences are subjected to random
mutagenesis. The resulting proteins are expressed, assessed for color, and the final color is determined as
described below.
Related Software: The Virtual Genetics Lab (VGLII) is a simulation of
genetics that allows more complex and varied models than Aipotu. Student solutions to problems in VGLII can be submitted
electronically for grading. Genetic models in VGLII are not elaborated at the DNA and protein levels as they are in Aipotu.
Biochemistry
In the Biochemistry Workbench, students use the tools of protein biochemistry to determine how
the colors of Aipotian flowers depend on the structure of their pigment proteins.
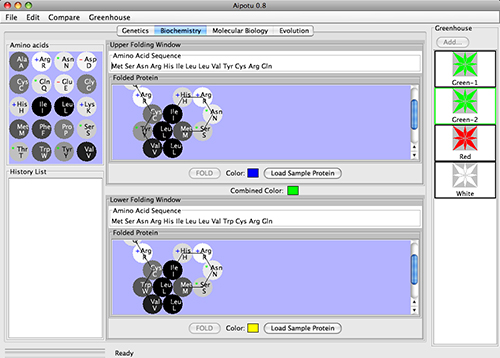
Students can:
- Examine the structures and colors of the pigment proteins found in any organism.
- Edit the amino acid sequences of existing proteins and observe the effect on their colors.
- Design proteins and observe their shapes and colors.
- Compare the amino acid sequences of different pigment proteins.
A record of each protein folded is preserved in the History List on the left side. Double-clicking an entry in the History List allows it to be brought to the main window so it can be explored in detail.
Algorithm: Proteins are folded on a 2-dimensional hexagonal lattice using an energy minimization
formula that includes ionic bonds, hydrogen bonds, and the hydrophobic interaction. Proteins with a particular
structure can be colored; the particular amino acids in that structure then determine the actual color.
Colored proteins combine to form overall colors via a specific set of rules.
Related Software: The Protein Investigator (PI) includes just the
protein folding software from Aipotu without the color determination. PI does include
the ability to fold in the presence or absence of disulfide bonds. PI also includes a "game mode" where users
design proteins to match particular target shapes; this mode can be used to provide graded homework problems.
Molecular Biology
In the Molecular Biology Workbench, students use the tools of molecular biology to determine how the different color alleles produce the color proteins.
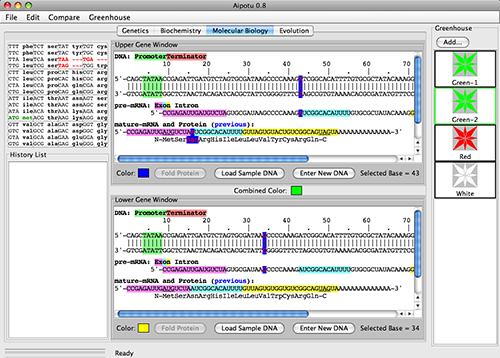
Students can:
- Examine the color-related DNA, mRNA, and protein sequences present in any organism.
- Click on any base in the DNA and see the corresponding bases in the pre-mRNA and mature mRNA as well as the amino acid in the protein.
- Edit the DNA sequence and observe the effect on RNA and protein sequences as well as the color of the protein produced.
- Compare DNA sequences of different alleles.
- Create organisms with DNA sequences they have designed and save them to the Greenhouse for exploration in the other Workbenches.
A record of each altered gene is preserved in the History List on the left side. Double-clickint an entry in the History List allows it to be brought to the main window so it can be explored in detail.
Algorithm: Aipotu searches the DNA sequence for a promoter (TATAA) and a terminator(GGGGG)
and then transcribes a pre-mRNA between them. It searches the pre-mRNA for intron start (GUGCG) and end
(CAAAG) sequences and processes the mRNA. It then searches for the 5'-most AUG and translates until a stop
codon or the 3' end of the mRNA is reached. The protein is then folded and its color is determined as
described in Biochemistry.
Related Software: The Gene Explorer (GX) includes just the
gene expression software from Aipotu without the color determination. GX also includes a "problem set mode" where users
design mutations to cause particular effects; this mode can be used to provide graded homework problems.
Evolution
In the Evolution Workbench, students use the tools of evolutionary analysis to explore the evolution of color in Aipotian flowers.
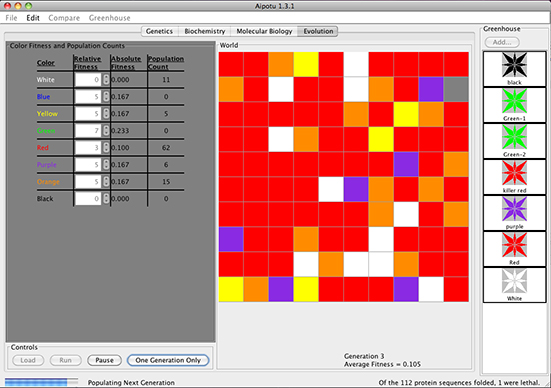
Students can:
- Load the World with organisms from the Greenhouse.
- Adjust the relative fitnesses of each color (even during a run).
- Run simulated evolution of Aipotian organisms.
- Extract evolved organisms from the World and save them to the Greenhouse for exploration in other Workbenches.
Algorithm: In each generation, each organism contributes copies of its two color gene DNA
sequences to the gene pool in proportion to its fitness. Each organism in the next generation is formed
by choosing two DNA sequences from the gene pool and subjecting them to random mutations at a user-settable
rate. Aipotu then uses the algorithm from Molecular Biology to determine the sequence
of the protein produced by each of the two DNA sequences. It then uses the algorithm from
Biochemistry to determine the color of each protein and the overall color of the organism.
These organisms are then displayed and the cycle repeats.







